Sen Zeng
Spatial Multi-Task Learning for Breast Cancer Molecular Subtype Prediction from Single-Phase DCE-MRI
Jan 11, 2026Abstract:Accurate molecular subtype classification is essential for personalized breast cancer treatment, yet conventional immunohistochemical analysis relies on invasive biopsies and is prone to sampling bias. Although dynamic contrast-enhanced magnetic resonance imaging (DCE-MRI) enables non-invasive tumor characterization, clinical workflows typically acquire only single-phase post-contrast images to reduce scan time and contrast agent dose. In this study, we propose a spatial multi-task learning framework for breast cancer molecular subtype prediction from clinically practical single-phase DCE-MRI. The framework simultaneously predicts estrogen receptor (ER), progesterone receptor (PR), human epidermal growth factor receptor 2 (HER2) status, and the Ki-67 proliferation index -- biomarkers that collectively define molecular subtypes. The architecture integrates a deep feature extraction network with multi-scale spatial attention to capture intratumoral and peritumoral characteristics, together with a region-of-interest weighting module that emphasizes the tumor core, rim, and surrounding tissue. Multi-task learning exploits biological correlations among biomarkers through shared representations with task-specific prediction branches. Experiments on a dataset of 960 cases (886 internal cases split 7:1:2 for training/validation/testing, and 74 external cases evaluated via five-fold cross-validation) demonstrate that the proposed method achieves an AUC of 0.893, 0.824, and 0.857 for ER, PR, and HER2 classification, respectively, and a mean absolute error of 8.2\% for Ki-67 regression, significantly outperforming radiomics and single-task deep learning baselines. These results indicate the feasibility of accurate, non-invasive molecular subtype prediction using standard imaging protocols.
TokenSeg: Efficient 3D Medical Image Segmentation via Hierarchical Visual Token Compression
Jan 08, 2026Abstract:Three-dimensional medical image segmentation is a fundamental yet computationally demanding task due to the cubic growth of voxel processing and the redundant computation on homogeneous regions. To address these limitations, we propose \textbf{TokenSeg}, a boundary-aware sparse token representation framework for efficient 3D medical volume segmentation. Specifically, (1) we design a \emph{multi-scale hierarchical encoder} that extracts 400 candidate tokens across four resolution levels to capture both global anatomical context and fine boundary details; (2) we introduce a \emph{boundary-aware tokenizer} that combines VQ-VAE quantization with importance scoring to select 100 salient tokens, over 60\% of which lie near tumor boundaries; and (3) we develop a \emph{sparse-to-dense decoder} that reconstructs full-resolution masks through token reprojection, progressive upsampling, and skip connections. Extensive experiments on a 3D breast DCE-MRI dataset comprising 960 cases demonstrate that TokenSeg achieves state-of-the-art performance with 94.49\% Dice and 89.61\% IoU, while reducing GPU memory and inference latency by 64\% and 68\%, respectively. To verify the generalization capability, our evaluations on MSD cardiac and brain MRI benchmark datasets demonstrate that TokenSeg consistently delivers optimal performance across heterogeneous anatomical structures. These results highlight the effectiveness of anatomically informed sparse representation for accurate and efficient 3D medical image segmentation.
Perspective+ Unet: Enhancing Segmentation with Bi-Path Fusion and Efficient Non-Local Attention for Superior Receptive Fields
Jun 20, 2024



Abstract:Precise segmentation of medical images is fundamental for extracting critical clinical information, which plays a pivotal role in enhancing the accuracy of diagnoses, formulating effective treatment plans, and improving patient outcomes. Although Convolutional Neural Networks (CNNs) and non-local attention methods have achieved notable success in medical image segmentation, they either struggle to capture long-range spatial dependencies due to their reliance on local features, or face significant computational and feature integration challenges when attempting to address this issue with global attention mechanisms. To overcome existing limitations in medical image segmentation, we propose a novel architecture, Perspective+ Unet. This framework is characterized by three major innovations: (i) It introduces a dual-pathway strategy at the encoder stage that combines the outcomes of traditional and dilated convolutions. This not only maintains the local receptive field but also significantly expands it, enabling better comprehension of the global structure of images while retaining detail sensitivity. (ii) The framework incorporates an efficient non-local transformer block, named ENLTB, which utilizes kernel function approximation for effective long-range dependency capture with linear computational and spatial complexity. (iii) A Spatial Cross-Scale Integrator strategy is employed to merge global dependencies and local contextual cues across model stages, meticulously refining features from various levels to harmonize global and local information. Experimental results on the ACDC and Synapse datasets demonstrate the effectiveness of our proposed Perspective+ Unet. The code is available in the supplementary material.
Neuro-Symbolic Learning: Principles and Applications in Ophthalmology
Jul 31, 2022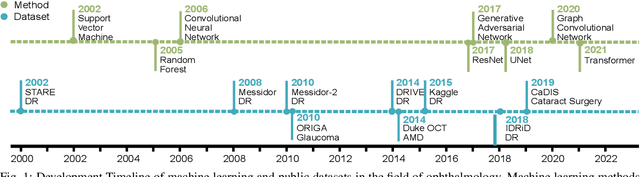
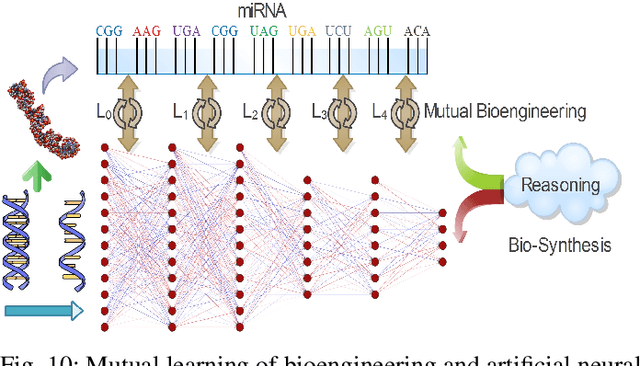
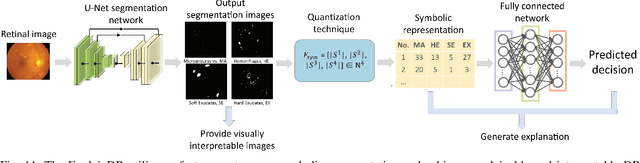
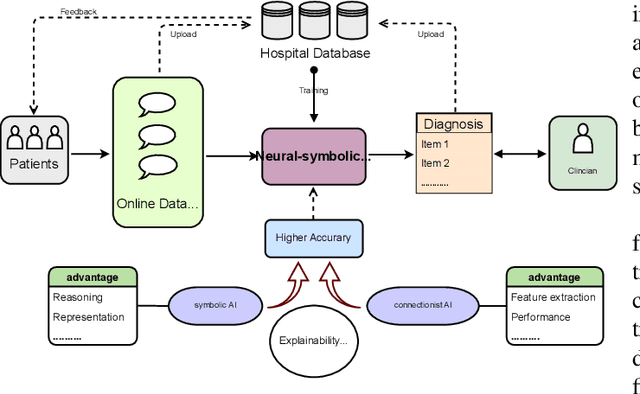
Abstract:Neural networks have been rapidly expanding in recent years, with novel strategies and applications. However, challenges such as interpretability, explainability, robustness, safety, trust, and sensibility remain unsolved in neural network technologies, despite the fact that they will unavoidably be addressed for critical applications. Attempts have been made to overcome the challenges in neural network computing by representing and embedding domain knowledge in terms of symbolic representations. Thus, the neuro-symbolic learning (NeSyL) notion emerged, which incorporates aspects of symbolic representation and bringing common sense into neural networks (NeSyL). In domains where interpretability, reasoning, and explainability are crucial, such as video and image captioning, question-answering and reasoning, health informatics, and genomics, NeSyL has shown promising outcomes. This review presents a comprehensive survey on the state-of-the-art NeSyL approaches, their principles, advances in machine and deep learning algorithms, applications such as opthalmology, and most importantly, future perspectives of this emerging field.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge